Q&A Report: Casting a Wider Net in Zebrafish Screening with Automated Microscopy and Image Analysis
Do you need expertise in machine learning to use the zebrafish detection software effectively?
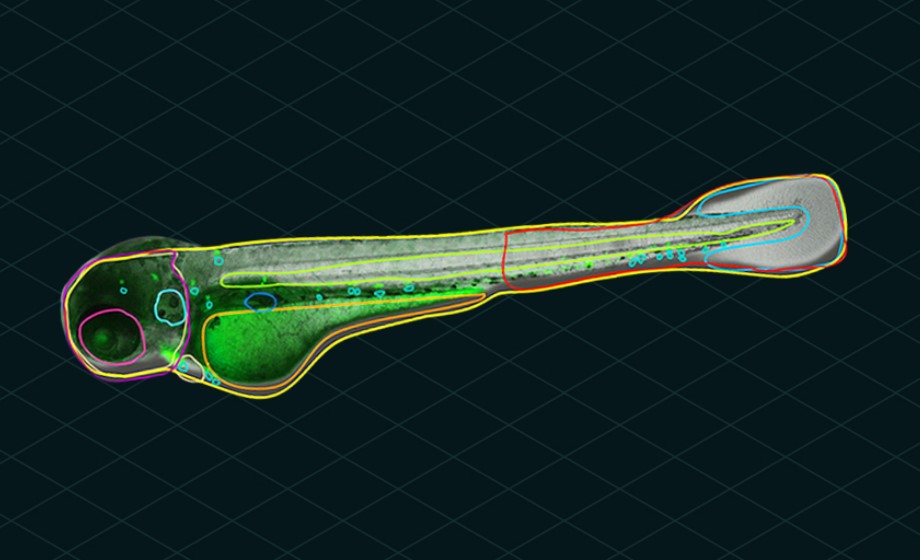
Our Athena image analysis software accompanying the WiScan® Hermes is designed to be straightforward and easy to use. Specifically, the AI-powered zebrafish analysis algorithm will automatically detect the zebrafish and internal organs without any required interaction from a user. The AI uses contrast information in the image to identify the fish and its anatomy all by itself. Once objects have been identified, then a user can optionally specify required area limits, thereby limiting the area of an image occupied by a fish or its anatomy to be above a minimum and/or below a maximum size.
Fluorescence images are analyzed in a standard fashion using a few parameters for removing background, edge detection and thresholding to identify bright objects. In this fashion, fluorescently labeled cells, organs and vasculature can be measured and studied in relation to the fish embryo’s underlying anatomy visible and detected in brightfield.
Can the software analyze similar data obtained from other platforms?
The Athena software can accept .tiff images for image analysis, both in brightfield and fluorescence. The images can be of 2D or 3D cell culture for analysis with our other analysis applications, or images of zebrafish for analysis with our AI-powered zebrafish application.
Beyond our existing Athena software, though, we are preparing to release a stand-alone version of our image analysis software in the near future. Through this platform, currently under development, zebrafish researchers will be able to analyze images acquired on other microscopes to identify the fish contour and internal anatomy in brightfield images in an unbiased and unsupervised fashion. As with our Athena application, structures identified in brightfield will be coupled to objects identified in fluorescence images. We aim to support images from multiple manufacturers and a variety of microscope types, including:
- Stereomicroscopes
- standard upright microscopes
- Non-Hermes inverted microscopes, both widefield and confocal
- Non-Hermes high-content screening (HCS) microscopes
Please contact IDEA Bio-Medical at info@idea-bio.com if you would like to be kept up to date regarding this software.
How flexible is the AI algorithm with different embryo phenotypes and different sample formats?
The Athena AI zebrafish application can be quite robust to changes in phenotype because it does not rely on external fiducial markers or have requirements for size, shape or morphology. So long as the morphology changes seen in the embryo maintain similar pixel contrast, then the software should be able to identify it well. We saw this in the recent article by Lubin et al., where they were able to identify microphthalmia in fish containing relevant mutations that cause a reduction in eye size. Because the eye of the fish maintained a similar darkness in the images for both wild type and mutant fish, the software was able to identify the eye with no additional user interaction. If a phenotype does happen to change the pixel intensity values when it is present, then additional AI training may be needed, but can be done quite easily and quickly.
What age of embryos are compatible with the AI?
So far, we have worked primarily with embryos between two and four days old and have had very good results. Embryos younger than two days are also supported and can be identified by the software, though their large yolk sac poses other challenges regarding sample preparation. Older embryos from five days onward should also be readily identified by the software, so long as they remain transparent. The difficulty in working with older embryos is that their swim bladder becomes more developed, causing the embryo to float within the sample.
What sample types and formats are supported?
For imaging, the Hermes will accept almost any sample type commonly used in biological laboratories, including 35-millimeter dishes and multi-well plates having anywhere from 6 to 1536 wells per plate. We have had quite good results come from the use of the 96 well alignment plate produced by the Hashimoto company from Japan [LINK], and from the use of standard 96 and 384 well plates with the fish embryos lying at the bottom of the well.
The advantage of using multi-well plates is obviously the accessible throughput. The Hermes microscope is able to scan a 96 well plate with one image per well in multiple colors in less than two minutes. Users of the system describe that if they perform Z stacks with five slices and four images per well in two colors in a 96 well plate, then typical scam times are between 15 to 17 minutes. Estimating approximately 20 minutes for image processing of a Z stack, followed by image analysis of another 20 minutes, the total time required to image and analyze one 96 well plate is about 45-60 minutes.