Q&A Report: Exploring Predictive Biomarkers and ERK1/2 Phosphorylation: A New Horizon in Forecasting Survival and Response to Immune Checkpoint Blockade in Glioblastoma Treatment
The answers to these questions have been provided by:
Víctor Andrés Arrieta, MD, PhD
Postdoctoral Fellow
Dept. of Neurosurgery
Northwestern University Feinberg School of Medicine
Can you comment about tumor heterogeneity and PD-1 blockade in glioblastoma? Can you comment on the value of multiplex immunofluorescence in studying tumor heterogeneity?
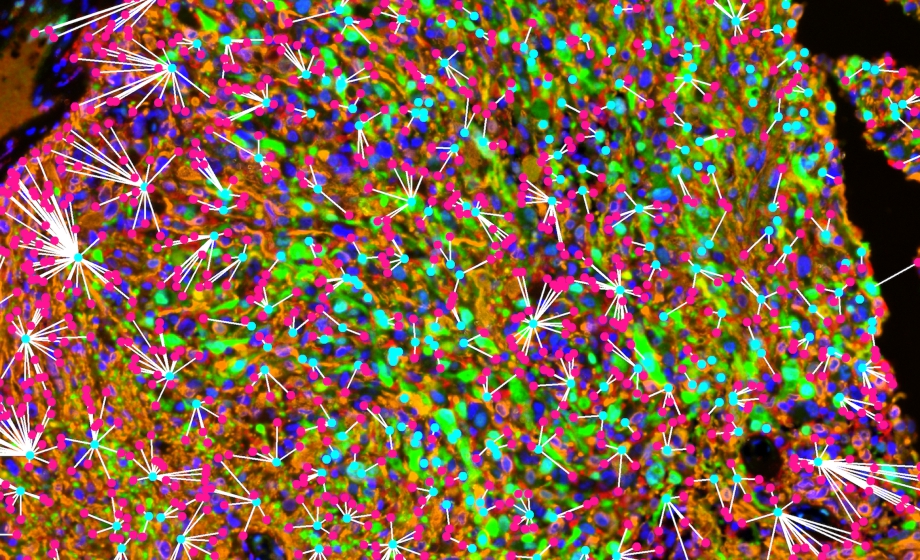
Glioblastoma is characterized by remarkable intra and intertumoral heterogeneity, both at the genetic and cellular levels. This heterogeneity can influence therapeutic responses, including to PD-1 blockade. Some regions of the tumor might be more immune-responsive than others, leading to varied outcomes with immunotherapy.
Multiplex immunofluorescence allows us to visualize multiple targets simultaneously within the tumor microenvironment, capturing the interactions between tumor cells, immune cells, and other stromal components. By doing so, we were able to gain insights into the spatial distribution and functional state of these cells, which can help in understanding the differential responses to therapies such as PD-1 blockade. With this, we were able to detect an interesting association of microglia with p-ERK in our clinical studies.
What might account for the varied responses to PD-1 blockade in glioblastoma patients, given that some show no difference while others experience prolonged survival?
That is an insightful question, highlighting the intricate nature of glioblastoma’s tumor microenvironment and the heterogeneity among patients. Drawing from our research and prior clinical trials, we discern that a significant clinical response is observed in only a minor fraction of GBM patients. Precise identification of these patients could potentially unlock significant therapeutic benefits. Several factors potentially determine the varied outcomes seen with PD-1 blockade in GBM patients:
- Patient’s Immune Health and Profile: The broader immunological health and history of a patient, encompassing both local and systemic immune functional status. We hypothesize that p-ERK levels might mirror the influence of the tumor’s immune milieu on the tumor phenotype. Reinforced by research spearheaded by Hara and Tirosh et al, it is evident that myeloid cells can, via cytokine secretion and other mechanisms, transform the glioma cell phenotype to be more receptive to immune recognition. Hence, the overarching strength and responsiveness of both local and systemic immune frameworks might sway not only the tumor cell’s nature but also the overarching efficacy of immunotherapies.
- Glioma Cellular Dynamics and Immune Interactions: We hypothesize that the specific cellular characteristics of gliomas and their interplay with immune cells are instrumental in dictating the efficacy of immunotherapy. For instance, certain immune entities, notably specific T cell subsets or myeloid cell groups, may either bolster or dampen the efficacy of PD-1 blockade.
Given the potential of p-ERK as a predictive biomarker, how do you see this influencing patient selection for anti-PD-1 therapy in the future, and what are the potential implications for improving outcomes in recurrent GBM patients?
Recognizing p-ERK as a potential predictive biomarker holds significant promise for the future of GBM treatment. Should further validation support its predictive capabilities, it offers a targeted approach to patient selection for anti-PD-1 therapy. Rather than employing a generalized ‘one-size-fits-all’ approach, we envision a more tailored therapeutic approach where GBM patients are chosen based on their p-ERK levels. Such a targeted strategy could amplify the effectiveness of anti-PD-1 therapy, holding the potential to elevate survival outcomes and enhance the quality of life for recurrent GBM patients.
Simultaneously, our ongoing project leveraging methylation data to ascertain the activation of the MAPK pathway underscores the progressive strides we are taking in this domain. The ubiquity of methylation profiling across numerous hospitals nationwide, combined with its ability to circumvent certain tissue preprocessing challenges inherent to immunohistochemistry, offers a robust dataset for analysis. Utilizing the latest in bioinformatics and computational methods, we anticipate that methylation profiling will further refine our ability to pinpoint those patients poised to gain the most from immunotherapies, heralding a new era of precision medicine in GBM treatment.
Are there other potential biomarkers that could be used to predict response to immunotherapy in GBM?
In the context of predicting response to immunotherapy in GBM, the most promising biomarkers currently identified include the activation of ERK1/2 cascade of the MAPK pathway, defects in the replication stress response in tumor cells, and germline POLE mutations. These three potential mechanisms have been associated with better survival in patients with GBM treated with anti-PD-1 therapy. While various other biomarkers, such as TMB and PD-L1 expression, have shown predictive potential in other cancers, their efficacy in predicting response in GBM remains uncertain. It is essential to highlight that, as of now, the predictability of many of these biomarkers in GBM is not as robust as in other malignancies. Further research is needed to validate these biomarkers and possibly identify new ones specific to GBM’s unique tumor microenvironment.
Did you use a standard multiplex immunofluorescence panel or have to create your own? What were the most critical factors you considered in developing a multiplex immunofluorescence panel for glioblastomas?
We developed our own multiplex immunofluorescence panel tailored for glioblastomas. The critical factors we considered for this panel included the clinical relevance of the markers that could inform about tumor biology in the context of p-ERK, the ability of this technology to capture the cellular heterogeneity of glioblastomas, the power to amplify fluorescent signals to accurately detect microglia, and the software development that allowed us to perform a spatial analysis of the cellular populations identified in the tumor microenvironment.
Did you perform the multiplex immunofluorescence on whole slides or specific regions of interest? How did you determine the intensity threshold to differentiate p-ERK positive from negative?
The multiplex immunofluorescence was done on specific regions of interest in the tumor that were chosen by the neuropathologist. With this, we established an intensity threshold using a tumor sample that had high expression of p-ERK to differentiate between p-ERK positive and negative.
What analysis software did you use for the multiplex immunofluorescence studies?
For the multiplex immunofluorescence studies, we primarily used inForm and Phenochart, software developed by Akoya. Once the data was processed with InForm, I conducted additional analyses using R.
How do you see the role of multiplex immunofluorescence and single-cell RNA sequencing evolving in future glioblastoma research, especially in relation to deciphering tumor-immune interactions?
Multiplex immunofluorescence (mIF) and single-cell RNA-seq (scRNA-seq) have emerged as pivotal tools in advancing our understanding of GBM. mIF sheds light on the spatial organization within the tumor, pinpointing where specific cell populations and proteins are situated in the tumor microenvironment. This spatial insight is crucial for grasping the intricate dynamics between tumor cells and the surrounding immune system. Complementing this, scRNA-seq delves into the intricate transcriptomic profiles of individual cells, offering a nuanced view of the diverse cellular states and functionalities present.
As we look to the future, I anticipate an even more synergistic application of these tools. The spatial clarity provided by mIF, when combined with the depth of scRNA-seq’s cell-by-cell analysis, has the potential to paint a holistic picture of tumor-immune interactions. Such integrated insights could be pivotal in unveiling novel therapeutic avenues and ensuring that patients receive treatments tailored to their unique tumor profiles.
Reflecting on recent clinical trials in GBM, while many have pursued positive clinical outcomes, a significant number lacked the depth of analysis needed to determine the optimal drug concentrations in the brain or to gauge the drug’s actual impact on both the tumor and its intricate immune milieu. With the advancing capabilities of mIF and scRNA-seq, I am optimistic that we will be better equipped to assess the true biological impact of experimental therapies on GBM. Furthermore, these technologies will be invaluable in dissecting the nuanced tumor-immune dynamics post-intervention, ensuring that our therapeutic strategies are always informed by the latest and most comprehensive data.